Running R at HCC
There are many methods to run R applications at HCC. I can break these uses down to:
- Creating a traditional Slurm submit file that runs an R script. The vast majority of R users do this.
- Using a program, such as GridR, that will create the submission files for you from within R.
In this post, I will discuss and layout the different methods of submitting jobs to HCC and the OSG. Further these methods lie on a spectrum of both difficulty in using.
 Each step is more and more difficult. Running R on your laptop is much easier than running R on a cluster. And running R on a cluster is less difficult than running it on the Grid. But there are techniques to bring these closer together.
Each step is more and more difficult. Running R on your laptop is much easier than running R on a cluster. And running R on a cluster is less difficult than running it on the Grid. But there are techniques to bring these closer together.
Creating Slurm submit files
Creating Slurm submit files and writing R scripts is the most common method of R users at HCC. The steps to this workflow is:
- Create a Slurm submit file
- Write a R script that will read in your data, and output it
- Copy data onto cluster from the laptop
- Submit Slurm submit file
- Wait for completion (you can ask to get an email)
More on the Slurm configuration is available at HCC Documentation.
A submit file for Slurm is below:
#!/bin/sh
#SBATCH --time=00:30:00
#SBATCH --mem-per-cpu=1024
#SBATCH --job-name=TestJob
#SBATCH --error=TestJob.stdout
#SBATCH --output=TestJob.stderr
module load R/3.3
R CMD BATCH Rcode.R
This submit file describes a job that will run 30 minutes, and require 1024MB of ram. Below the #SBATCH lines are the actual script that will run on the worker node. The module command loads the newest version of R on HCC’s clusters. Next command runs an R script named Rcode.R.
A parallel submission is:
#!/bin/sh
#SBATCH --ntasks-per-node=16
#SBATCH --nodes=1
#SBATCH --time=00:30:00
#SBATCH --mem-per-cpu=1024
#SBATCH --job-name=TestJob
#SBATCH --error=TestJob.stdout
#SBATCH --output=TestJob.stderr
module load R/3.3
R CMD BATCH Rcode.R
This submit file adds --ntasks-per-node and --nodes=1 that describes the parallel jobs. ntasks-per-node specifies how many cores on a remote worker node is required for the job. --nodes describes the number of physical nodes that this job should span across. All other lines are very similar to the previous single core submission file.
The R code looks a bit different though. Here is an example:
library(“parallel")
a <- function(s) { return (2*s) }
mclapply(c(1:20), a, mc.cores = 16)
This will run mclapply which will apply the made up function a across the list specified in c(1, 20).
Using GridR to submit processing
GridR is another method for farming processing out to remote cluster. GridR is able to submit to HTCondor clusters. Therefore, it is able to submit to the OSG through HTCondor.
The GridR package is hosted on Github. The wiki is very useful with examples and tutorials on how to use GridR.
Below is a working example script of using R on HCC’s Crane cluster.
library(GridR)
# Initialize the GridR library for submissions
grid.init(service="condor.local", localTmpDir="tmp", bootstrap=TRUE, remoteRPath="/util/opt/R/3.3/gcc/4.4/bin/R", Rurl="https://www.dropbox.com/s/s27ngq1rp7e9qeb/el6-R-modified_1.0.tar.gz?dl=0")
# Create a quick function to run remotely
a <- function(s) { return (2*s) }
# Run the apply function, much like lapply. In this case, with only 1 attribute to apply
grid.apply("x", a, 13, wait = TRUE)
# Output the results.
x
This R script submits jobs to the OSG from the Crane cluster. It will run the simple a function on the remote worker nodes on the OSG.
The jobs can run anywhere on the OSG:
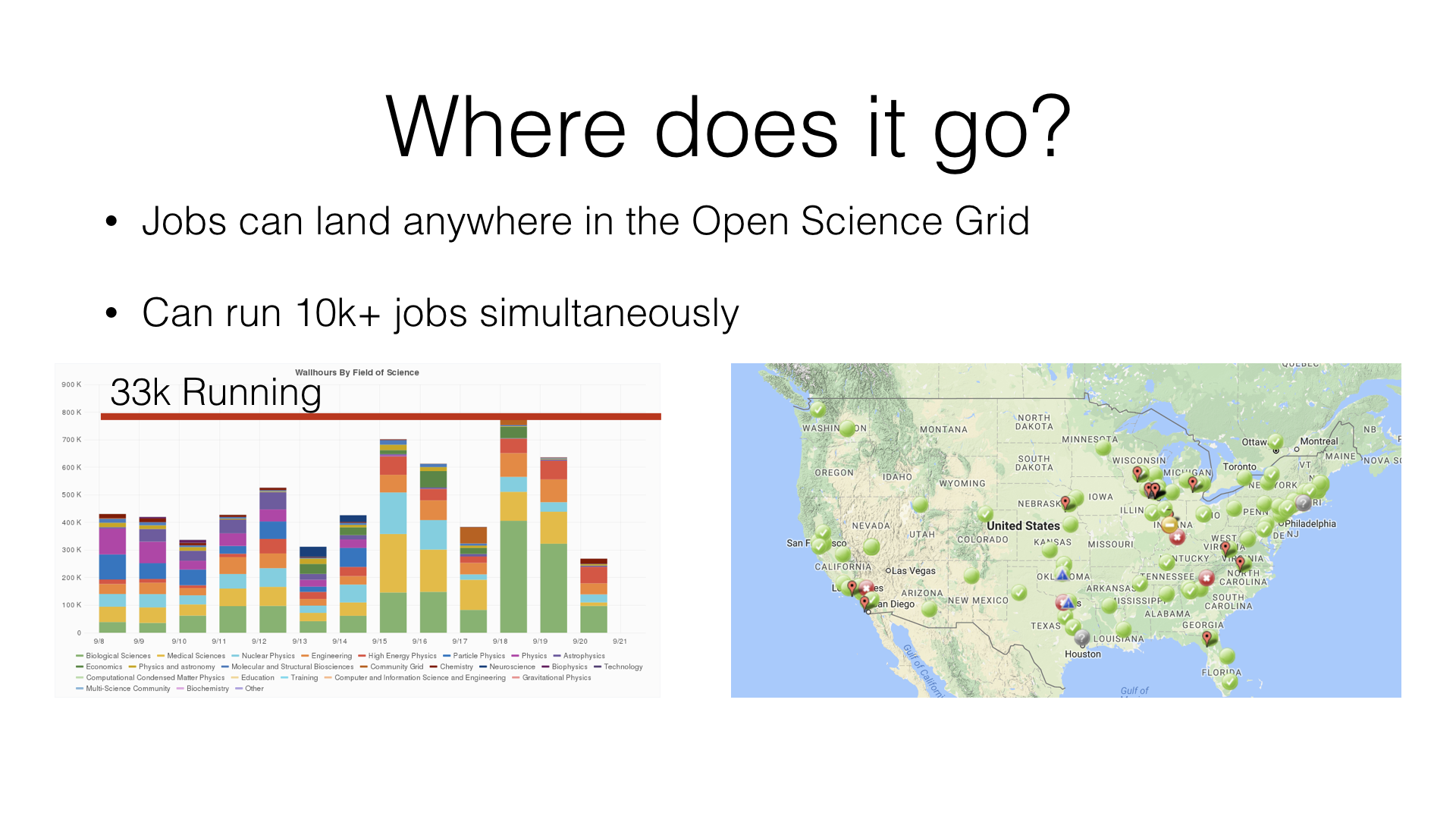
Jobs submitted to the OSG can run on multiple sites around the U.S. They will execute and and return the results.
Conclusion
There are many methods to submitting R processing to clusters and the grid. One has to choose which one best suites them.
The GridR method is easy for experience R programmers. But, it lacks the flexibility of the Slurm submit file method. The Slurm submit method requires learning some Linux and Slurm syntax, but offers the flexibility to specify multiple cores per R script or more memory per job.


Leave a comment